NA-MIC Project Weeks
NA-MIC Project Weeks
Back to Projects List
Evaluation of ModelHub.ai
Key Investigators
- Hans Meine (Uni Bremen, Fraunhofer MEVIS)
Project Description
Much like the recent integration of NVIDIA’s AI-assisted annotation in Slicer and various other platforms, there’s the more vendor-neutral http://modelhub.ai (originated at Harvard MS / BWH / Data-Farber). This project seems to be very well thought-through and documented, and recently got interesting models as well.
Objective
- Try out running models from http://modelhub.ai
- Possibly integrate in MeVisLab
- Compare with AIAA and other solutions
Approach and Plan
- Install and run modelhub.ai software
- Investigate which models are interesting (e.g. liver & tumor segmentation)
- Try running models
- Find out how to integrate in MeVisLab or Slicer
Progress and Next Steps
- Taking a closer look, modelhub.ai seems to be very well-designed, but got less traction and less models than AIAA
- The API includes sample data for each model, making it trivial to test whether they work.
- The API links models to publications, licenses for the model, the sample data, and modelhub itself.
- However, there is only sparse technical metadata, compared with AIAA.
- The website allows to browse models (much more convenient than NVIDIA’s GPU cloud).
- However, there are not many interesting medical imaging ones.
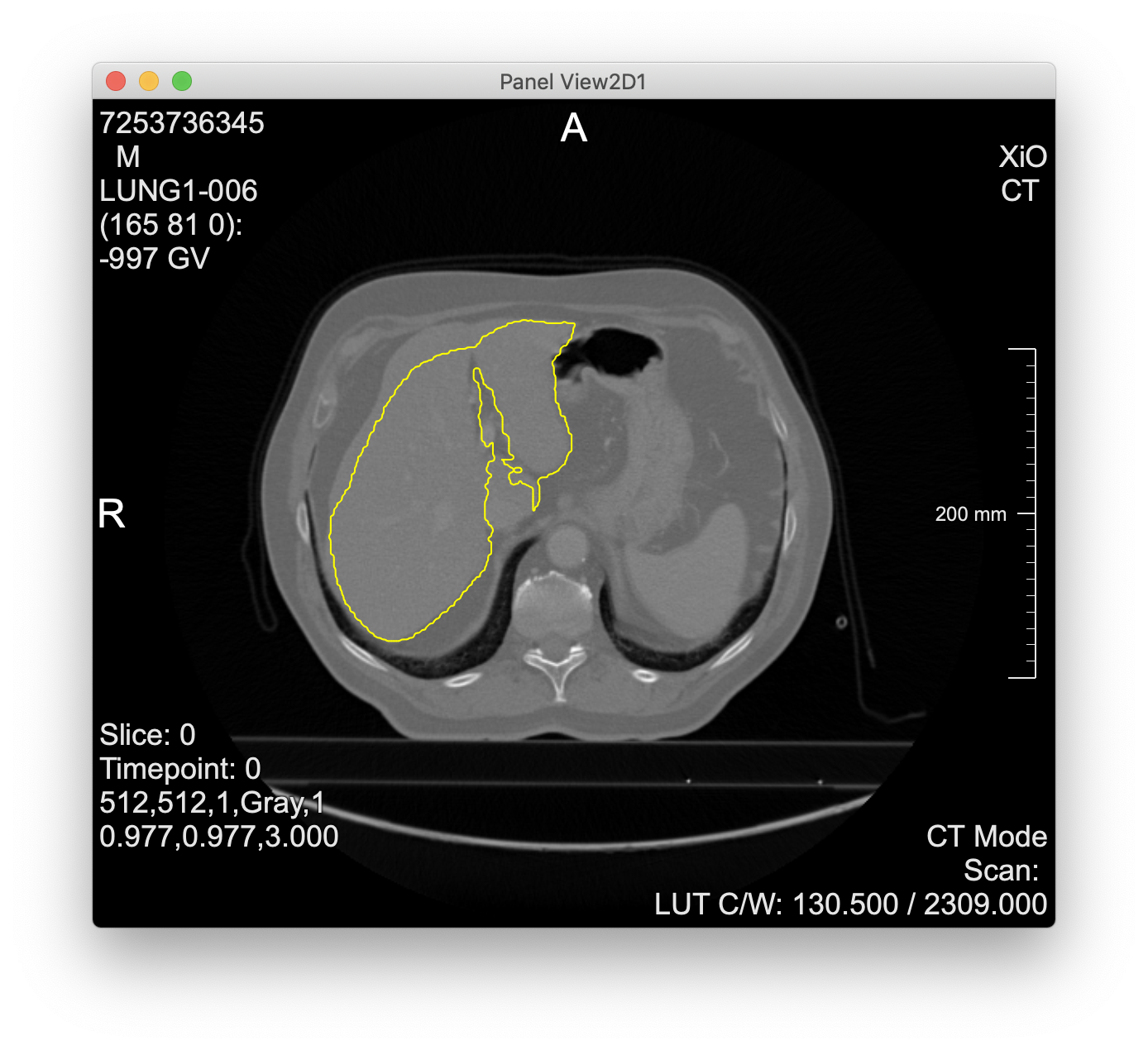
- cascaded-fcn-liver is an interesting model (from the organizers of the LiTS challenge)
- deep-prognosis gives a 2-year survival prognosis based on a NSCLC tumor ROI
- Evaluation of cascaded-fcn-liver
- takes a single CT slice as DICOM file via a form-encoded POST request
- returns contours as voxel coordinates in JSON format
- screenshot from MeVisLab experiments below
- “cascaded” = two networks for liver + tumor segmentation, but the API runs only the first (the second is included, but execution is left to the user)
- Evaluation of deep-prognosis
- takes a 150x150x150 numpy array file in .npy format (again as form-encoded POST request)
- the linked paper mentioned a 50x50x50 ROI, so there was a discrepancy, but the API gave clear requirements before running the model and a clear error message when feeding input of the wrong size
- Conclusions on http://modelhub.ai
- was really not much work to get running
- not many models available today, but nice open platform
Illustrations
Liver contours computed via the cascaded-fcn-liver model parsed and visualized in MeVisLab: