 NA-MIC Project Weeks
NA-MIC Project Weeks
Back to Projects List
Real-time visualization in transcranial magnetic stimulation (TMS)
Key Investigators
- Loraine Franke (University of Massachusetts Boston)
- Lipeng Ning (BWH & Harvard Medical School)
- Yogesh Rathi (BWH & Harvard Medical School)
- Steve Pieper (Isomics, Inc.)
- Daniel Haehn (University of Massachusetts Boston)
Project Description
Transcranial magnetic stimulation is a nonivasive procedure used for treating depression with magnetic and electric fields to stimulate nerve cells. A TMS coil is slowly moved over the subject’s head suface to target certain areas in the brain. Our project aims to develop a deep-learning powered software for real-time E-Field prediction and a visualization of TMS within 3D Slicer.
Objective
Real-time visualization of an electric field (E-field) for transcranial magnetic stimulation (TMS) on the brain surface as well as visualizing the E-field along fiber bundles (DTI), and later the possibility to optimize the exact coil position or to target certain fibers.
Approach and Plan
Integrate the visualization process as a new module within the MRML scene architecture:
- Evaluate different methods to visualize the E-field on the surface mesh (find fastest method which is appropriate for a real-time visualization)
- Rendering of a virtual TMS coil in the 3D Slicer module
- Develop an approach to move/rotate the coil model over the brain surface in 3D Slicer (similar to markups/fiducials)
Progress and Next Steps
- Visualize Efield (volume) on brain surface (polydata/mesh) by adjusting the models orientation and applying the vtkProbeFilter within a new Slicer Module.
- Tested the rendering time of our visualization approach with renderer.GetLastRenderTimeInSeconds() resulted in an average of 0.8 milliseconds.
- Added a functionality to create fiducials and to move it along the brain model’s surface.
Next steps:
- Replace the fiducial with a TMS coil model.
- Compare with further methods for rendering time (vtkpointlocator, cppyy, manual with optimized storage)
- Apply vector field visualization on tractography data / pick fibers (similar to DBS Navigation ).
Illustrations
Fiducial (yellow sphere) moving along the brain surface with mapped vector field:
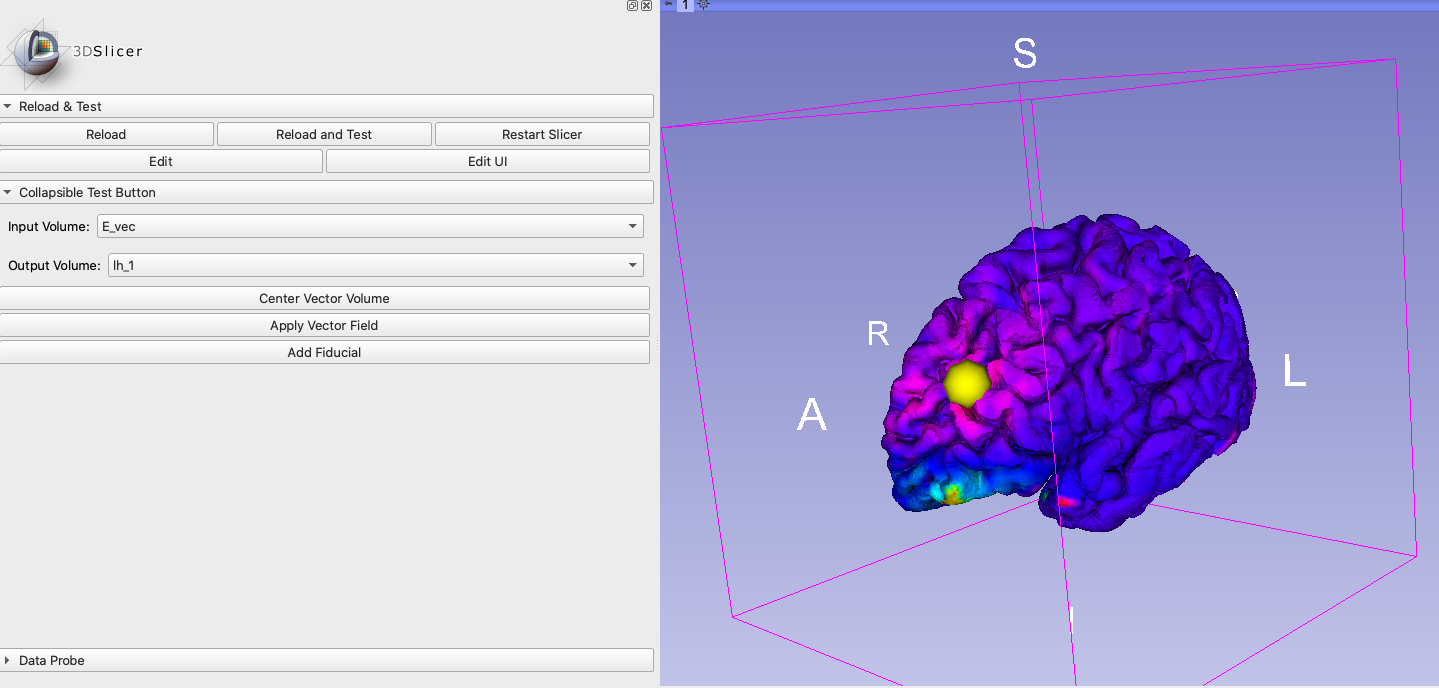 The fiducial in this screenshot will later be replaced by a TMS coil model.
The fiducial in this screenshot will later be replaced by a TMS coil model.
The vector field volume and the brain surface mesh overlapping after applying the module’s functionalities:
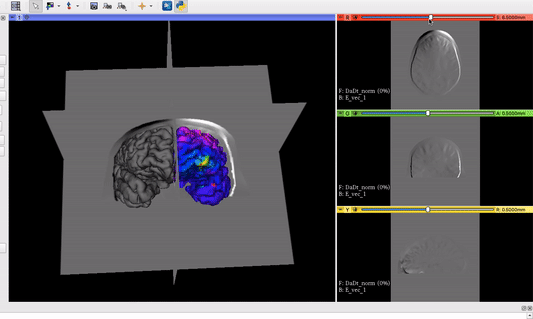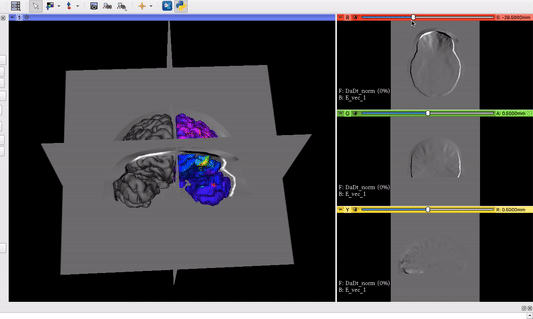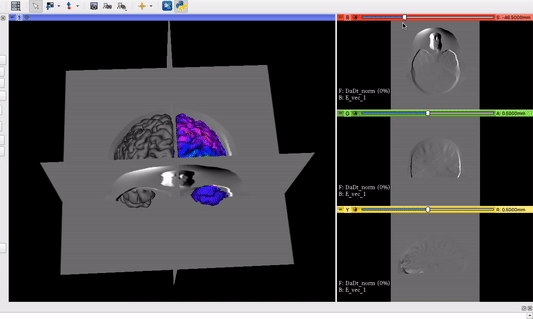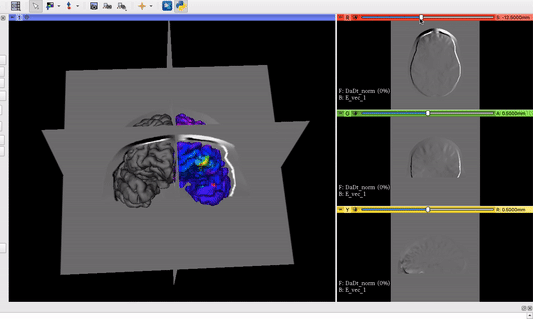
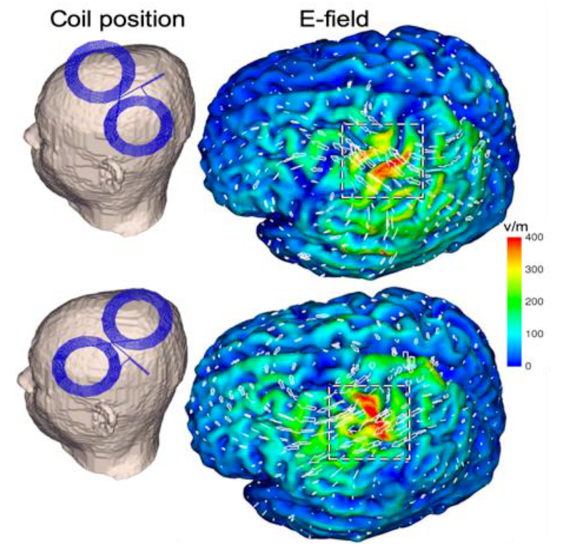
Visualization goal from another software we want to implement as Module in 3D Slicer:
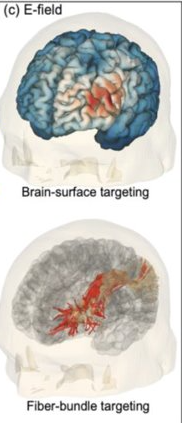
Visualization process also on tractography for fiber bundle targeting:
Background and References
vtkProbeFilter: https://vtk.org/doc/nightly/html/classvtkProbeFilter.html Moving fiducials with CPYY: https://gist.github.com/pieper/f9da3e0a73c70981b48d0747132526d5
Measure rendering time in 3D Slicer:
- Getting renderer: https://slicer.readthedocs.io/en/latest/developer_guide/script_repository.html#access-vtk-views-renderers-and-cameras
- Then applying renderer.GetLastRenderTimeInSeconds()
- Random note following the project presentations: https://cyclotronresearchcentre.github.io/forward/ might be useful for using diffusion-weighted imaging to compute conductivity tensors in the white matter to make the electromagnetic simulations more accurate.